概要
breast cancerデータはUCIの機械学習リポジトリ―にあるBreast Cancer Wisconsin (Diagnostic) Data Setのコピーで、乳腺腫瘤の穿刺吸引細胞診(fine needle aspirate (FNA) of a breast mass)のデジタル画像から計算されたデータ。
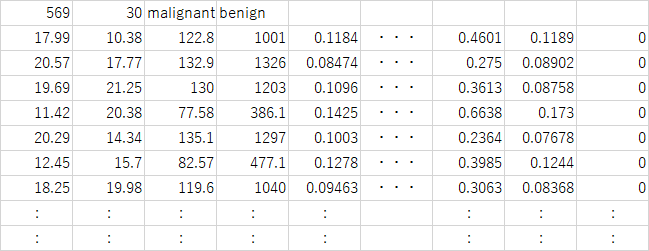
乳癌に関する細胞テストの様々な数値と、その結果のデータセット。569人の被検者の複数の腫瘤に関する細胞診の結果得られた30個の特徴量と、各被験者の診断結果(悪性/良性:benign/malignant)が格納されている。
ここではPythonのscikit-learnにあるbreast_cancerデータの使い方をまとめる。
データの取得とデータ構造
Pythonで扱う場合、scikit-learnのdatasetsモジュールにあるload_breast_cancer()でデータを取得できる。データはBunchクラスのオブジェクト。
|
1 2 3 4 5 6 |
from sklearn.datasets import load_breast_cancer cancer_ds = load_breast_cancer() for key, value in zip(cancer_ds.keys(), cancer_ds.values()): print("{}:\n{}\n".format(key, value)) |
データの構造は辞書型で、569人の細胞診の結果に関する30個の特徴量をレコードとしたの配列、各腫瘤の診断結果など。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 |
data: [[1.799e+01 1.038e+01 1.228e+02 ... 2.654e-01 4.601e-01 1.189e-01] [2.057e+01 1.777e+01 1.329e+02 ... 1.860e-01 2.750e-01 8.902e-02] [1.969e+01 2.125e+01 1.300e+02 ... 2.430e-01 3.613e-01 8.758e-02] ... [1.660e+01 2.808e+01 1.083e+02 ... 1.418e-01 2.218e-01 7.820e-02] [2.060e+01 2.933e+01 1.401e+02 ... 2.650e-01 4.087e-01 1.240e-01] [7.760e+00 2.454e+01 4.792e+01 ... 0.000e+00 2.871e-01 7.039e-02]] target: [0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 1 0 1 1 1 1 1 0 0 1 0 0 1 1 1 1 0 1 0 0 1 1 1 1 0 1 0 0 1 0 1 0 0 1 1 1 0 0 1 0 0 0 1 1 1 0 1 1 0 0 1 1 1 0 0 1 1 1 1 0 1 1 0 1 1 ..... 1 1 1 1 1 1 0 1 0 1 1 0 1 1 1 1 1 0 0 1 0 1 0 1 1 1 1 1 0 1 1 0 1 0 1 0 0 1 1 1 0 1 1 1 1 1 1 1 1 1 1 1 0 1 0 0 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 0 0 0 0 0 0 1] target_names: ['malignant' 'benign'] DESCR: .. _breast_cancer_dataset: Breast cancer wisconsin (diagnostic) dataset -------------------------------------------- **Data Set Characteristics:** :Number of Instances: 569 :Number of Attributes: 30 numeric, predictive attributes and the class :Attribute Information: - radius (mean of distances from center to points on the perimeter) - texture (standard deviation of gray-scale values) - perimeter - area - smoothness (local variation in radius lengths) - compactness (perimeter^2 / area - 1.0) - concavity (severity of concave portions of the contour) - concave points (number of concave portions of the contour) - symmetry - fractal dimension ("coastline approximation" - 1) The mean, standard error, and "worst" or largest (mean of the three largest values) of these features were computed for each image, resulting in 30 features. For instance, field 3 is Mean Radius, field 13 is Radius SE, field 23 is Worst Radius. - class: - WDBC-Malignant - WDBC-Benign :Summary Statistics: ===================================== ====== ====== Min Max ===================================== ====== ====== radius (mean): 6.981 28.11 texture (mean): 9.71 39.28 perimeter (mean): 43.79 188.5 area (mean): 143.5 2501.0 smoothness (mean): 0.053 0.163 compactness (mean): 0.019 0.345 concavity (mean): 0.0 0.427 concave points (mean): 0.0 0.201 symmetry (mean): 0.106 0.304 fractal dimension (mean): 0.05 0.097 radius (standard error): 0.112 2.873 texture (standard error): 0.36 4.885 perimeter (standard error): 0.757 21.98 area (standard error): 6.802 542.2 smoothness (standard error): 0.002 0.031 compactness (standard error): 0.002 0.135 concavity (standard error): 0.0 0.396 concave points (standard error): 0.0 0.053 symmetry (standard error): 0.008 0.079 fractal dimension (standard error): 0.001 0.03 radius (worst): 7.93 36.04 texture (worst): 12.02 49.54 perimeter (worst): 50.41 251.2 area (worst): 185.2 4254.0 smoothness (worst): 0.071 0.223 compactness (worst): 0.027 1.058 concavity (worst): 0.0 1.252 concave points (worst): 0.0 0.291 symmetry (worst): 0.156 0.664 fractal dimension (worst): 0.055 0.208 ===================================== ====== ====== :Missing Attribute Values: None :Class Distribution: 212 - Malignant, 357 - Benign :Creator: Dr. William H. Wolberg, W. Nick Street, Olvi L. Mangasarian :Donor: Nick Street :Date: November, 1995 This is a copy of UCI ML Breast Cancer Wisconsin (Diagnostic) datasets. https://goo.gl/U2Uwz2 Features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. They describe characteristics of the cell nuclei present in the image. Separating plane described above was obtained using Multisurface Method-Tree (MSM-T) [K. P. Bennett, "Decision Tree Construction Via Linear Programming." Proceedings of the 4th Midwest Artificial Intelligence and Cognitive Science Society, pp. 97-101, 1992], a classification method which uses linear programming to construct a decision tree. Relevant features were selected using an exhaustive search in the space of 1-4 features and 1-3 separating planes. The actual linear program used to obtain the separating plane in the 3-dimensional space is that described in: [K. P. Bennett and O. L. Mangasarian: "Robust Linear Programming Discrimination of Two Linearly Inseparable Sets", Optimization Methods and Software 1, 1992, 23-34]. This database is also available through the UW CS ftp server: ftp ftp.cs.wisc.edu cd math-prog/cpo-dataset/machine-learn/WDBC/ .. topic:: References - W.N. Street, W.H. Wolberg and O.L. Mangasarian. Nuclear feature extraction for breast tumor diagnosis. IS&T/SPIE 1993 International Symposium on Electronic Imaging: Science and Technology, volume 1905, pages 861-870, San Jose, CA, 1993. - O.L. Mangasarian, W.N. Street and W.H. Wolberg. Breast cancer diagnosis and prognosis via linear programming. Operations Research, 43(4), pages 570-577, July-August 1995. - W.H. Wolberg, W.N. Street, and O.L. Mangasarian. Machine learning techniques to diagnose breast cancer from fine-needle aspirates. Cancer Letters 77 (1994) 163-171. feature_names: ['mean radius' 'mean texture' 'mean perimeter' 'mean area' 'mean smoothness' 'mean compactness' 'mean concavity' 'mean concave points' 'mean symmetry' 'mean fractal dimension' 'radius error' 'texture error' 'perimeter error' 'area error' 'smoothness error' 'compactness error' 'concavity error' 'concave points error' 'symmetry error' 'fractal dimension error' 'worst radius' 'worst texture' 'worst perimeter' 'worst area' 'worst smoothness' 'worst compactness' 'worst concavity' 'worst concave points' 'worst symmetry' 'worst fractal dimension'] filename: C:\Users\tomo\AppData\Local\Programs\Python\Python37-32\lib\site-packages\sklearn\datasets\data\breast_cancer.csv |
データのキーは以下のようになっている。
|
1 2 3 4 5 6 7 |
from sklearn.datasets import load_breast_cancer cancer_ds = load_breast_cancer() print(cancer_ds.keys()) # dict_keys(['data', 'target', 'target_names', 'DESCR', 'feature_names', 'filename']) |
データの内容
'data'~特徴量データセット
569人の細胞診結果に対する30個の特徴量のデータを格納した2次元配列。列のインデックス(0~29)が30個の特徴量に対応している。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 |
'data': array([[1.799e+01, 1.038e+01, 1.228e+02, ..., 2.654e-01, 4.601e-01, 1.189e-01], [2.057e+01, 1.777e+01, 1.329e+02, ..., 1.860e-01, 2.750e-01, 8.902e-02], [1.969e+01, 2.125e+01, 1.300e+02, ..., 2.430e-01, 3.613e-01, 8.758e-02], ..., [1.660e+01, 2.808e+01, 1.083e+02, ..., 1.418e-01, 2.218e-01, 7.820e-02], [2.060e+01, 2.933e+01, 1.401e+02, ..., 2.650e-01, 4.087e-01, 1.240e-01], [7.760e+00, 2.454e+01, 4.792e+01, ..., 0.000e+00, 2.871e-01, 7.039e-02]]) |
'target'~診断結果に対応したコード
各被検者の診断結果(悪性:malignant、良性:benign)を格納した0/1のコードの配列。569個の腫瘤に対応した1次元配列(0:悪性が212、1:良性が357)。
|
1 2 3 4 5 6 7 |
'target': array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 1, 1, 0, 1, 0, 0, ..... 1, 1, 1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 1]) |
'target_names'~診断結果
診断結果、悪性(malignant)/良性(benign)が定義されている。。
|
1 |
'target_names': array(['malignant', 'benign'], dtype='<U9') |
診断結果とコードの関係は以下の通り。
| malignant | 0 |
| benign | 1 |
'feature_names'~特徴名
データの格納順はDESCRの後。細胞診の結果得られた30個の特徴量の名前。
腫瘤に関する以下の10の属性について、それぞれ平均(mean)、標準偏差(error)、最悪値(worst)の3種類、合計30の特性値に対する名前が格納されている。ここでworstは各属性に関する最大値となっている。
- radius:半径(中心から外周までの平均)
- texture:テクスチャ―のグレースケールの標準偏差
- perimeter:外周長
- area:面積
- smoothness:中心から外周までの部分偏差
- compactness:コンパクト性(外周長2÷面積-1.0)
- concavity:コンターの凹部強度
- concave points:コンターの凹点の数
- symmetry:対称性
- fractal dimension:フラクタル次元
|
1 2 3 4 5 6 7 8 9 10 |
'feature_names': array(['mean radius', 'mean texture', 'mean perimeter', 'mean area', 'mean smoothness', 'mean compactness', 'mean concavity', 'mean concave points', 'mean symmetry', 'mean fractal dimension', 'radius error', 'texture error', 'perimeter error', 'area error', 'smoothness error', 'compactness error', 'concavity error', 'concave points error', 'symmetry error', 'fractal dimension error', 'worst radius', 'worst texture', 'worst perimeter', 'worst area', 'worst smoothness', 'worst compactness', 'worst concavity', 'worst concave points', 'worst symmetry', 'worst fractal dimension'], dtype='<U23') |
特徴名とコードの関係は以下の通り。
| mean | error | worst | |
| radius | 0 | 10 | 20 |
| texture | 1 | 11 | 21 |
| perimeter | 2 | 12 | 22 |
| area | 3 | 13 | 23 |
| smoothness | 4 | 14 | 24 |
| compactness | 5 | 15 | 25 |
| concavity | 6 | 16 | 26 |
| concave points | 7 | 17 | 27 |
| symmetry | 8 | 18 | 28 |
| fractal dimension | 9 | 19 | 29 |
'filename'~ファイル名
これも格納順はDESCRの後で、CSVファイルの位置が示されている。1行目にはデータ数、特徴量数、特徴量名称が並んでおり、その後に569行のレコードに対する4列の特徴量と1列の診断結果データが格納されている。このファイルにはfeature_namesやDESCRに当たるデータは格納されていない。
|
1 |
'filename': 'C:...\\lib\\site-packages\\sklearn\\datasets\\data\\breast_cancer.csv' |
'DESCR'~データセットの説明
データセットの説明。print(breast_ds_dataset['DESCR'])のようにprint文で整形表示される。
- レコード数569個(悪性:212、良性:357)
- 属性は、30の数値属性とクラス
→predictiveの意味とclassが単数形なのがわからない
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 |
.. _breast_cancer_dataset: Breast cancer wisconsin (diagnostic) dataset -------------------------------------------- **Data Set Characteristics:** :Number of Instances: 569 :Number of Attributes: 30 numeric, predictive attributes and the class :Attribute Information: - radius (mean of distances from center to points on the perimeter) - texture (standard deviation of gray-scale values) - perimeter - area - smoothness (local variation in radius lengths) - compactness (perimeter^2 / area - 1.0) - concavity (severity of concave portions of the contour) - concave points (number of concave portions of the contour) - symmetry - fractal dimension ("coastline approximation" - 1) The mean, standard error, and "worst" or largest (mean of the three largest values) of these features were computed for each image, resulting in 30 features. For instance, field 3 is Mean Radius, field 13 is Radius SE, field 23 is Worst Radius. - class: - WDBC-Malignant - WDBC-Benign :Summary Statistics: ===================================== ====== ====== Min Max ===================================== ====== ====== radius (mean): 6.981 28.11 texture (mean): 9.71 39.28 perimeter (mean): 43.79 188.5 area (mean): 143.5 2501.0 smoothness (mean): 0.053 0.163 compactness (mean): 0.019 0.345 concavity (mean): 0.0 0.427 concave points (mean): 0.0 0.201 symmetry (mean): 0.106 0.304 fractal dimension (mean): 0.05 0.097 radius (standard error): 0.112 2.873 texture (standard error): 0.36 4.885 perimeter (standard error): 0.757 21.98 area (standard error): 6.802 542.2 smoothness (standard error): 0.002 0.031 compactness (standard error): 0.002 0.135 concavity (standard error): 0.0 0.396 concave points (standard error): 0.0 0.053 symmetry (standard error): 0.008 0.079 fractal dimension (standard error): 0.001 0.03 radius (worst): 7.93 36.04 texture (worst): 12.02 49.54 perimeter (worst): 50.41 251.2 area (worst): 185.2 4254.0 smoothness (worst): 0.071 0.223 compactness (worst): 0.027 1.058 concavity (worst): 0.0 1.252 concave points (worst): 0.0 0.291 symmetry (worst): 0.156 0.664 fractal dimension (worst): 0.055 0.208 ===================================== ====== ====== :Missing Attribute Values: None :Class Distribution: 212 - Malignant, 357 - Benign :Creator: Dr. William H. Wolberg, W. Nick Street, Olvi L. Mangasarian :Donor: Nick Street :Date: November, 1995 This is a copy of UCI ML Breast Cancer Wisconsin (Diagnostic) datasets. https://goo.gl/U2Uwz2 Features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. They describe characteristics of the cell nuclei present in the image. Separating plane described above was obtained using Multisurface Method-Tree (MSM-T) [K. P. Bennett, "Decision Tree Construction Via Linear Programming." Proceedings of the 4th Midwest Artificial Intelligence and Cognitive Science Society, pp. 97-101, 1992], a classification method which uses linear programming to construct a decision tree. Relevant features were selected using an exhaustive search in the space of 1-4 features and 1-3 separating planes. The actual linear program used to obtain the separating plane in the 3-dimensional space is that described in: [K. P. Bennett and O. L. Mangasarian: "Robust Linear Programming Discrimination of Two Linearly Inseparable Sets", Optimization Methods and Software 1, 1992, 23-34]. This database is also available through the UW CS ftp server: ftp ftp.cs.wisc.edu cd math-prog/cpo-dataset/machine-learn/WDBC/ .. topic:: References - W.N. Street, W.H. Wolberg and O.L. Mangasarian. Nuclear feature extraction for breast tumor diagnosis. IS&T/SPIE 1993 International Symposium on Electronic Imaging: Science and Technology, volume 1905, pages 861-870, San Jose, CA, 1993. - O.L. Mangasarian, W.N. Street and W.H. Wolberg. Breast cancer diagnosis and prognosis via linear programming. Operations Research, 43(4), pages 570-577, July-August 1995. - W.H. Wolberg, W.N. Street, and O.L. Mangasarian. Machine learning techniques to diagnose breast cancer from fine-needle aspirates. Cancer Letters 77 (1994) 163-171. |
データの利用
データの取得方法
breast_cancerデータセットから各データを取り出すのに、以下の2つの方法がある。
- 辞書のキーを使って呼び出す(例:
breast_cancer_dataset['DESCR']) - キーの文字列をプロパティーに指定する(例:
breast_cancer_dataset.DESCR)
全レコードの特徴量データの取得
'data'から、569のレコードに関する30の特徴量が569行30列の2次元配列で得られる。30の特徴量は'feature_names'の30の特徴名に対応している。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 |
from sklearn.datasets import load_breast_cancer cancer_ds = load_breast_cancer() print(cancer_ds.data) # [[1.799e+01 1.038e+01 1.228e+02 ... 2.654e-01 4.601e-01 1.189e-01] # [2.057e+01 1.777e+01 1.329e+02 ... 1.860e-01 2.750e-01 8.902e-02] # [1.969e+01 2.125e+01 1.300e+02 ... 2.430e-01 3.613e-01 8.758e-02] # ... # [1.660e+01 2.808e+01 1.083e+02 ... 1.418e-01 2.218e-01 7.820e-02] # [2.060e+01 2.933e+01 1.401e+02 ... 2.650e-01 4.087e-01 1.240e-01] # [7.760e+00 2.454e+01 4.792e+01 ... 0.000e+00 2.871e-01 7.039e-02]] |
特定の特徴量のデータのみ取得
特定の特徴量に関する全レコードのデータを取り出すときにはX[:, n]の形で指定する。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 |
from sklearn.datasets import load_breast_cancer cancer_ds = load_breast_cancer() features = cancer_ds['feature_names'] X = cancer_ds['data'] n_feature = 10 feature = X[:, n_feature] print("feature name : {}".format(features[n_feature])) print("feature data :\n{}".format(feature)) # feature name : radius error # feature data : # [1.095 0.5435 0.7456 0.4956 0.7572 0.3345 0.4467 0.5835 0.3063 0.2976 # 0.3795 0.5058 0.9555 0.4033 0.2121 0.37 0.4727 0.5692 0.7582 0.2699 # 0.1852 0.2773 0.4388 0.6917 0.8068 1.046 0.2545 0.8529 0.439 0.6003 # ..... # 0.2784 0.2542 0.3031 0.2351 0.272 0.346 0.2104 0.1144 0.2957 0.5196 # 0.3163 0.28 0.2409 0.3013 0.2116 0.2199 0.2441 0.5375 0.2254 0.2388 # 0.3645 0.3141 0.2602 0.9622 1.176 0.7655 0.4564 0.726 0.3857] |
特定のクラスのデータのみ抽出
特定のクラス(この場合は診断結果)のレコードのみを抽出する方法。ndarrayの条件による要素抽出を使う。
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 |
from sklearn.datasets import load_breast_cancer cancer_ds = load_breast_cancer() targets = cancer_ds['target_names'] X = cancer_ds['data'] y = cancer_ds['target'] n_class = 0 data_0 = X[:, n_class] print("data for class {}:\n{}".format(targets[n_class], X[y==n_class])) # data for class malignant: # [[1.799e+01 1.038e+01 1.228e+02 ... 2.654e-01 4.601e-01 1.189e-01] # [2.057e+01 1.777e+01 1.329e+02 ... 1.860e-01 2.750e-01 8.902e-02] # [1.969e+01 2.125e+01 1.300e+02 ... 2.430e-01 3.613e-01 8.758e-02] # ... # [2.013e+01 2.825e+01 1.312e+02 ... 1.628e-01 2.572e-01 6.637e-02] # [1.660e+01 2.808e+01 1.083e+02 ... 1.418e-01 2.218e-01 7.820e-02] [2.060e+01 2.933e+01 1.401e+02 ... 2.650e-01 4.087e-01 1.240e-01]] |